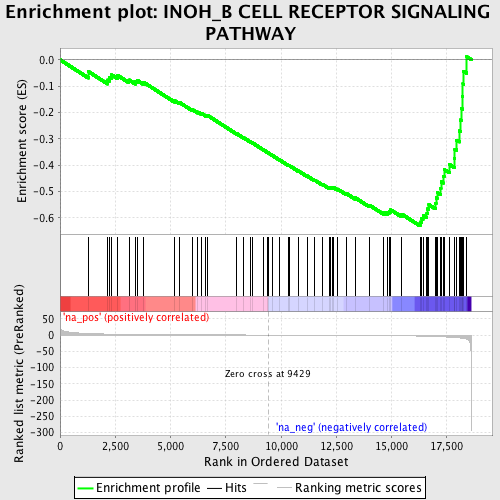
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | INOH_B CELL RECEPTOR SIGNALING PATHWAY |
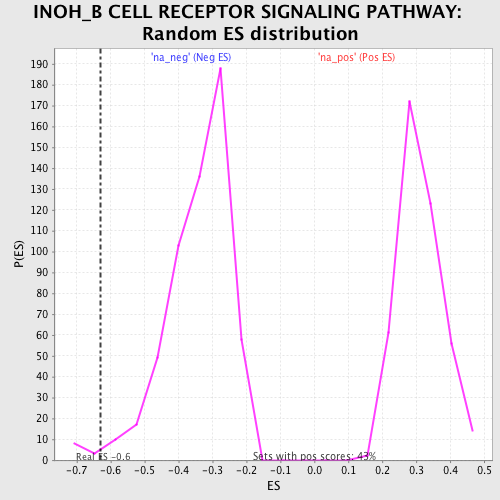
| Enrichment Score (ES) | -0.6304568 |
| Normalized Enrichment Score (NES) | -1.8364061 |
| Nominal p-value | 0.015734266 |
| FDR q-value | 0.50633 |
| FWER p-Value | 0.985 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TRAF4 | 1281 | 4.890 | -0.0447 | No | ||
| 2 | GRB7 | 2157 | 3.067 | -0.0765 | No | ||
| 3 | TRAF3 | 2235 | 2.939 | -0.0660 | No | ||
| 4 | CRK | 2332 | 2.817 | -0.0571 | No | ||
| 5 | PRKCZ | 2606 | 2.511 | -0.0593 | No | ||
| 6 | MYD88 | 3117 | 2.089 | -0.0764 | No | ||
| 7 | MAP2K1 | 3408 | 1.872 | -0.0827 | No | ||
| 8 | ITPR3 | 3484 | 1.824 | -0.0776 | No | ||
| 9 | FRS3 | 3768 | 1.671 | -0.0846 | No | ||
| 10 | SRC | 5173 | 1.116 | -0.1547 | No | ||
| 11 | MAP2K2 | 5396 | 1.043 | -0.1615 | No | ||
| 12 | GRAP2 | 5998 | 0.867 | -0.1895 | No | ||
| 13 | CRADD | 6229 | 0.806 | -0.1979 | No | ||
| 14 | PRKCE | 6403 | 0.763 | -0.2034 | No | ||
| 15 | SHC3 | 6599 | 0.710 | -0.2104 | No | ||
| 16 | GRB10 | 6685 | 0.688 | -0.2115 | No | ||
| 17 | PIK3R2 | 7997 | 0.358 | -0.2804 | No | ||
| 18 | ITPR1 | 8317 | 0.274 | -0.2963 | No | ||
| 19 | DOK2 | 8633 | 0.201 | -0.3122 | No | ||
| 20 | BCAR1 | 8716 | 0.180 | -0.3158 | No | ||
| 21 | HRAS | 9216 | 0.053 | -0.3424 | No | ||
| 22 | PRKCI | 9371 | 0.014 | -0.3506 | No | ||
| 23 | LAT | 9409 | 0.005 | -0.3526 | No | ||
| 24 | NRAS | 9596 | -0.046 | -0.3624 | No | ||
| 25 | TRAF2 | 9913 | -0.132 | -0.3788 | No | ||
| 26 | SOS1 | 10324 | -0.226 | -0.3998 | No | ||
| 27 | CBL | 10381 | -0.236 | -0.4016 | No | ||
| 28 | PRKCH | 10782 | -0.328 | -0.4215 | No | ||
| 29 | ARAF | 11175 | -0.429 | -0.4405 | No | ||
| 30 | NFKBIB | 11524 | -0.516 | -0.4567 | No | ||
| 31 | PIK3R1 | 11865 | -0.608 | -0.4720 | No | ||
| 32 | TRAF1 | 12200 | -0.703 | -0.4865 | No | ||
| 33 | TRAF5 | 12232 | -0.711 | -0.4846 | No | ||
| 34 | PRKCQ | 12320 | -0.741 | -0.4856 | No | ||
| 35 | IRS1 | 12354 | -0.755 | -0.4836 | No | ||
| 36 | FADD | 12551 | -0.815 | -0.4901 | No | ||
| 37 | GAB2 | 12943 | -0.928 | -0.5066 | No | ||
| 38 | IRS4 | 13356 | -1.072 | -0.5235 | No | ||
| 39 | GRB14 | 13992 | -1.318 | -0.5511 | No | ||
| 40 | PRKCD | 14653 | -1.601 | -0.5787 | No | ||
| 41 | RAF1 | 14806 | -1.685 | -0.5785 | No | ||
| 42 | SHC1 | 14889 | -1.736 | -0.5743 | No | ||
| 43 | CD19 | 14965 | -1.777 | -0.5694 | No | ||
| 44 | LCP2 | 15453 | -2.130 | -0.5851 | No | ||
| 45 | FYN | 16296 | -3.029 | -0.6153 | Yes | ||
| 46 | SOS2 | 16367 | -3.158 | -0.6034 | Yes | ||
| 47 | PIK3CA | 16426 | -3.246 | -0.5903 | Yes | ||
| 48 | PIK3CD | 16600 | -3.558 | -0.5819 | Yes | ||
| 49 | GRB2 | 16640 | -3.636 | -0.5658 | Yes | ||
| 50 | CD79A | 16691 | -3.727 | -0.5499 | Yes | ||
| 51 | LCK | 16984 | -4.337 | -0.5441 | Yes | ||
| 52 | BLNK | 17018 | -4.436 | -0.5237 | Yes | ||
| 53 | RELA | 17065 | -4.586 | -0.5033 | Yes | ||
| 54 | CRKL | 17222 | -5.001 | -0.4868 | Yes | ||
| 55 | SH2D2A | 17241 | -5.040 | -0.4626 | Yes | ||
| 56 | MAPK3 | 17335 | -5.344 | -0.4409 | Yes | ||
| 57 | NFKB1 | 17383 | -5.487 | -0.4161 | Yes | ||
| 58 | NFKBIA | 17625 | -6.306 | -0.3976 | Yes | ||
| 59 | NFKBIE | 17840 | -7.112 | -0.3737 | Yes | ||
| 60 | DOK1 | 17867 | -7.248 | -0.3389 | Yes | ||
| 61 | SH2B3 | 17944 | -7.663 | -0.3048 | Yes | ||
| 62 | SYK | 18076 | -8.451 | -0.2697 | Yes | ||
| 63 | VAV1 | 18125 | -8.843 | -0.2282 | Yes | ||
| 64 | PRKCA | 18153 | -9.054 | -0.1845 | Yes | ||
| 65 | CD79B | 18213 | -9.597 | -0.1398 | Yes | ||
| 66 | TRAF6 | 18232 | -9.851 | -0.0916 | Yes | ||
| 67 | GAB1 | 18256 | -10.039 | -0.0427 | Yes | ||
| 68 | KRAS | 18407 | -12.449 | 0.0113 | Yes |